Github中文镜像_github镜像 知乎
Hi,大家好,我是编程小6,很荣幸遇见你,我把这些年在开发过程中遇到的问题或想法写出来,今天说一说Github中文镜像_github镜像 知乎,希望能够帮助你!!!。
血清型分析网站
Center for Genomic Epidemiology
1.介绍
血清型又称血清蛋白型,指存在于血浆中蛋白质的遗传标记。
血清型是指病毒和细菌中特定不同的亚种。这些微生物一般都由细胞表层的抗原来分类命名的。但是在同一种类的微生物里也会有分支和不同。
血清型的决定是由很多种类的因素而形成的,其中不乏包括病毒性、革兰氏阴性菌表层的脂多糖、外毒素、细菌的质粒和噬菌体等。通过聚合酶链式反应,也可以利用基因特性来分别不同的血清型。
一般来说,血清分型是基于体细胞O抗原 (somatic O antigen)、细胞表面蛋白和构成鞭毛一部分的H抗原或基于荚膜多糖(CPS)K抗原
2.沙门菌血清型预测
H+O,测试使用GCF_000006945.2
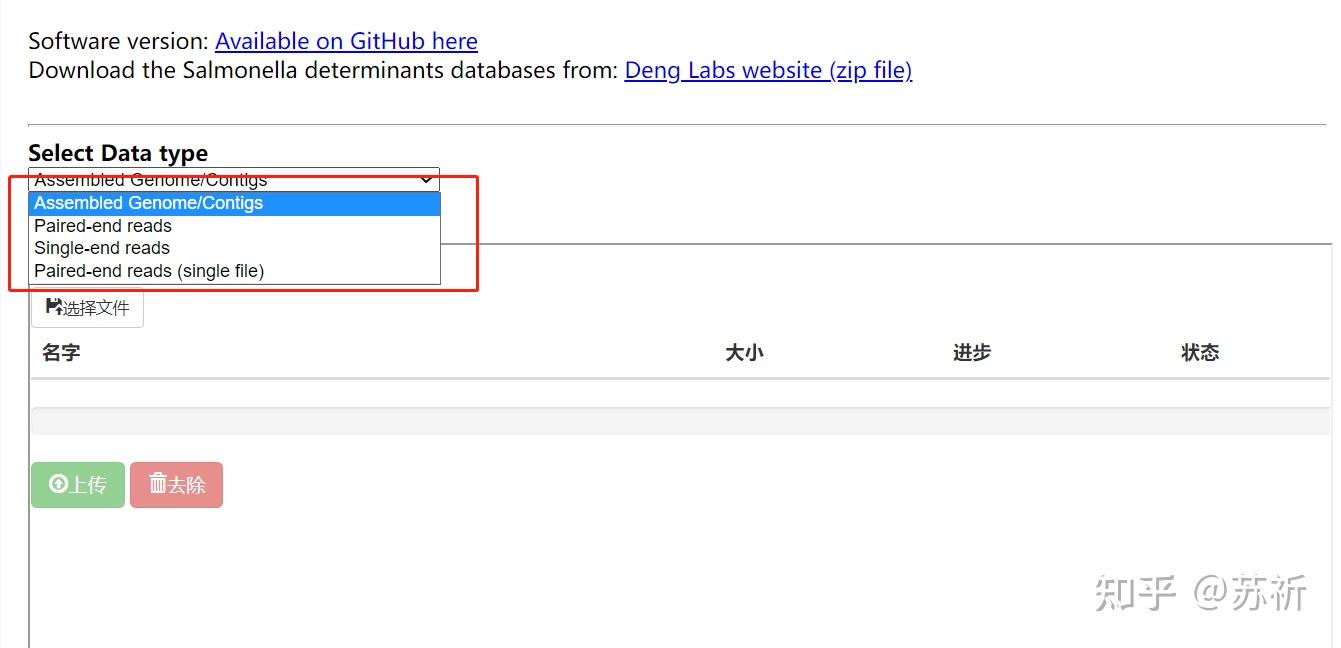
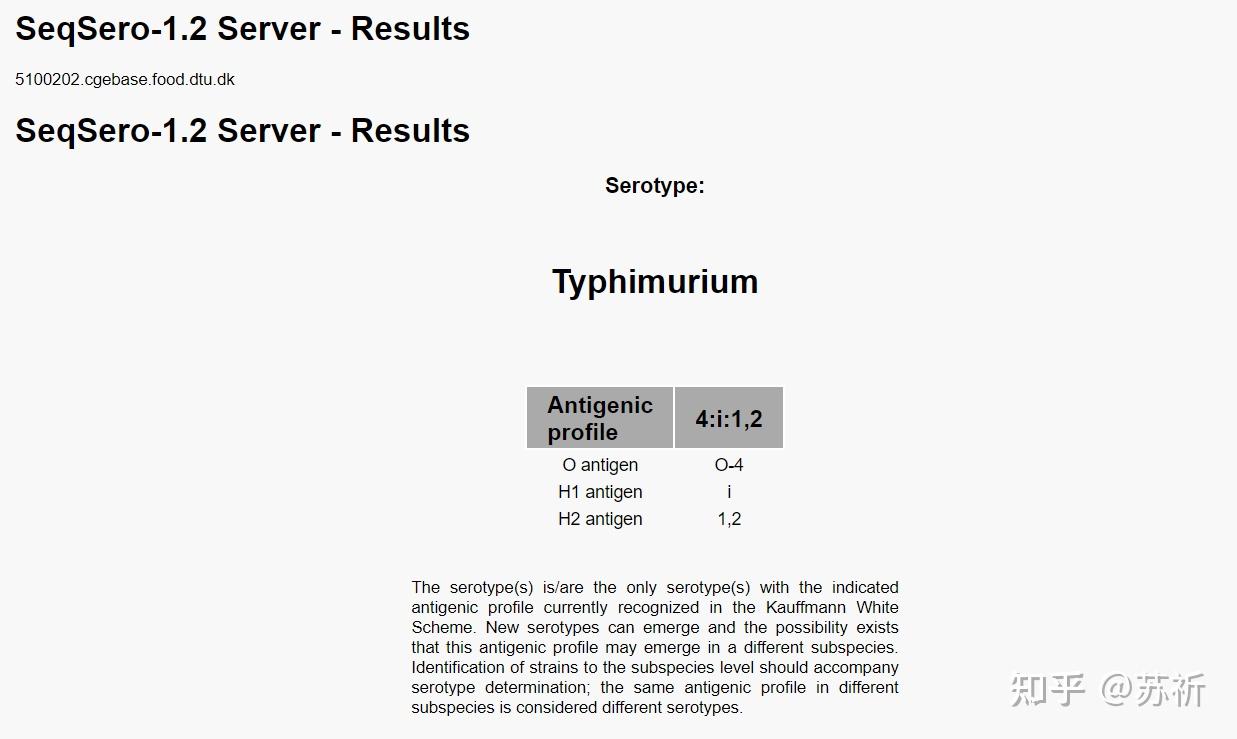
2.1 SeqSero,后续版本SeqSero2
SeqSero:https://cge.food.dtu.dk/services/SeqSero/
SeqSero2:SeqSero-Salmonella Serotyping by Whole Genome Sequencing
支持使用原始reads数据或者全基因组组装序列进行分析,
2.2 MOST血清型预测
英国公共卫生部开发的MOST工具,利用SRST工具分析全基因组测序数据,并结合MLST分型结果,以识别相应的血清型
https://github.com/phe-bioinformatics/MOST
2.3 SalmonellaTypeFinder
丹麦科技大学开发的网页版工具SalmonellaTypeFinder,运行SeqSero预测血清型并应用SRST2确定MLST分型,最终结合两种分型工具的结果预测沙门菌的血清型
Bitbucket
在线网站链接好像失效了
2.4 SISTR
SISTR则是结合cgMLST分型策略预测沙门菌的血清型,其准确率可达94.6%
https://github.com/phac-nml/sistr_cmd#web-application
3.大肠埃希菌血清型预测
大肠埃希菌的菌体抗原(O)、荚膜抗原(K)、鞭毛抗原(H)和菌毛抗原(F)是其血清型鉴定和分型的基础
测试使用GCF_000005845.2
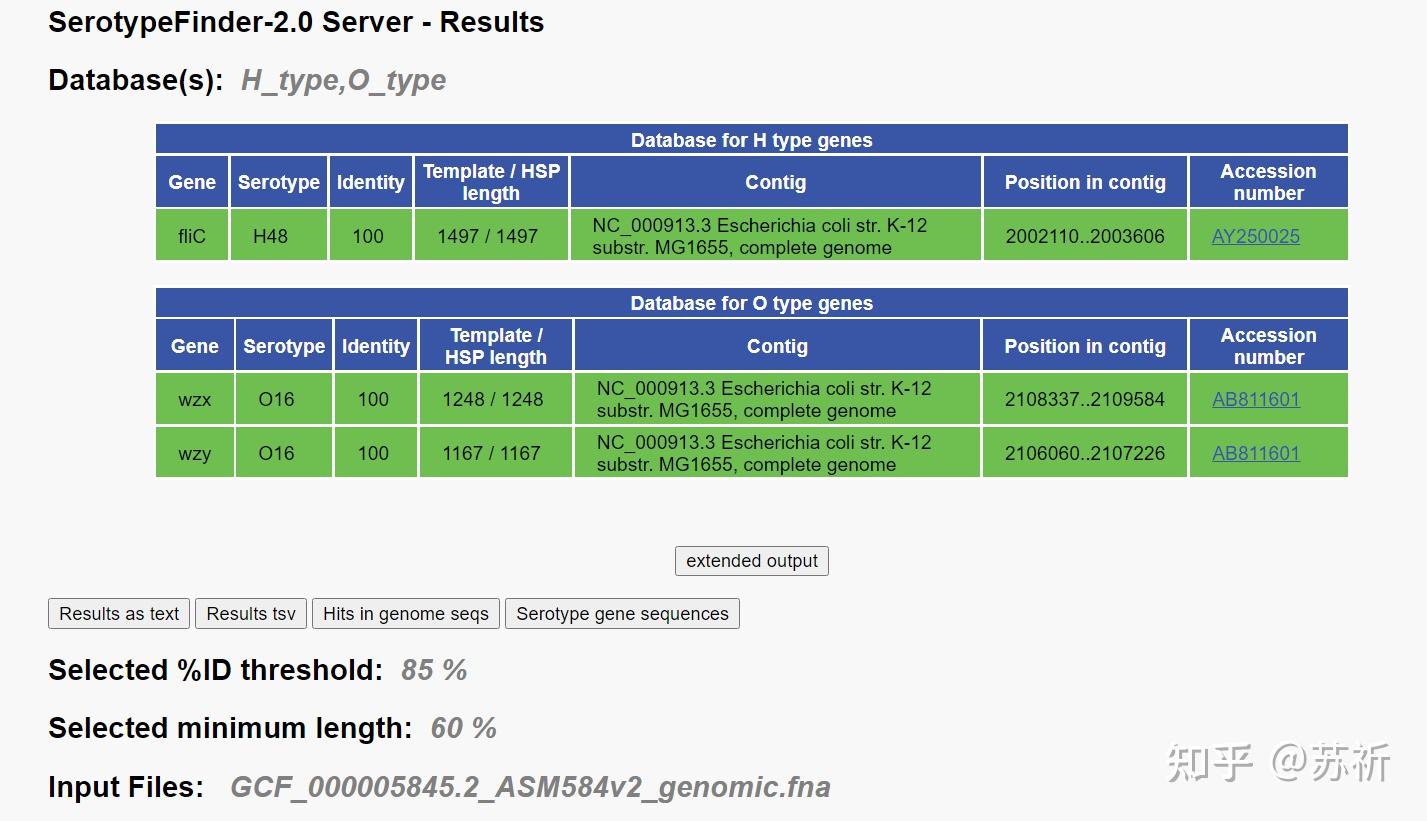
3.1 SerotypeFinder
Joensen等学者创建基于大肠埃希菌全基因组数据预测其血清型分型的在线分析工具SerotypeFinder,该工具基于特定O抗原基因(wzx、wzy、wzm和wzt)和H抗原基因(fliC、flkA、fllA、flmA和flnA)序列的分型方法与传统血清学结果具有高度一致性
3.2 ClermonTyping
在线工具ClermonTyping可将大肠埃希菌细分为A、B1、B2、C、D、E和F七个主要的亚群(phylogroups),从而替代传统的多重PCR方法对大肠埃希菌进行亚种分型
http://clermontyping.iame-research.center

4.肺炎克雷伯菌血清型预测
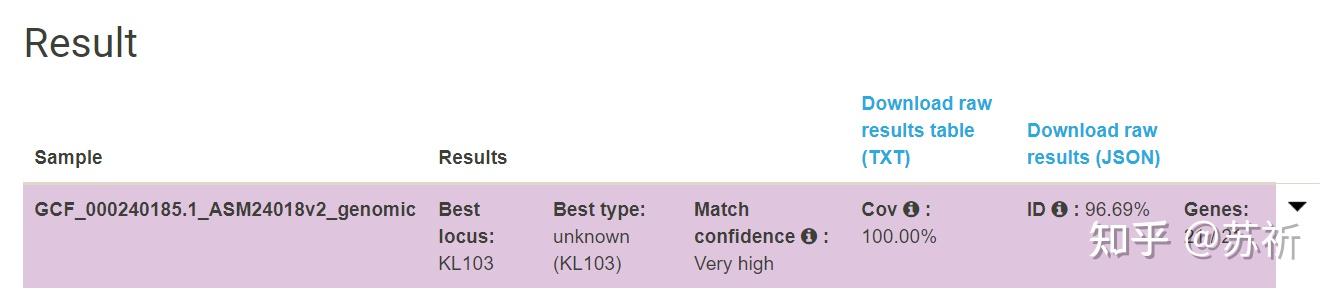
K+O,测试使用GCF_000240185.1
4.1 Kaptive
Kaptive Web是一种快速鉴别肺炎克雷伯菌K (编码荚膜多糖)和O位点(编码脂多糖O抗原)的在线工具


4.2 Pathogenwatch
pathogen.watch在线网站也可以做肺炎克雷伯菌的血清型预测,预测操作见:Kleborate:肺炎克雷伯菌分析

5.鲍曼不动杆菌
2020年最新版本的Kaptive Web增加了对鲍曼不动杆菌荚膜多糖(capsular polysaccharide,KL)和脂寡糖外核心(lipooligosaccharide outer core,OCL)的识别
表格:
| Name | Description | Accessibility | Link |
| chewBBACA | cg/wgMLST schema creation and allele calling using genome assemblies (FASTA) |
Web & Standalone | https://chewbbaca.online https://github.com/B-UMMI/chewBBACA |
| MentaLiST | cg/wgMLST schema creation and allele calling using raw sequencing reads (FASTQ) or genome assemblies (FASTA) |
Standalone | https://github.com/WGS-TB/MentaLiST |
| Genome profiler | cg/wgMLST schema creation and allele calling using genome assemblies (FASTA) |
Standalone | http://sourceforge.net/projects/genomeprofiler |
| In silico typing | |||
| SeqSero | Salmonella serotype prediction using raw sequencing reads (FASTQ) or genome assemblies (FASTA) |
Web & Standalone | http://denglab.info/SeqSero https://github.com/denglab/SeqSero |
| SeqSero2 | Similar to SeqSero (an updated version) | Web & Standalone | https://github.com/denglab/SeqSero2 http://www.denglab.info/SeqSero2 |
| MOST | Salmonella serotype prediction and MLST allele calling using raw sequencing reads (FASTQ) |
Standalone | https://github.com/phe-bioinformatics/MOST |
| SalmonellaTypeFinder | Salmonella serotype prediction and MLST allele calling using raw sequencing reads (FASTQ) |
Web & Standalone | https://bitbucket.org/genomicepidemiology/salmonellatypefinder/src/master/ https://cge.cbs.dtu.dk/services/SalmonellaTypeFinder |
| SISTR | Salmonella serotype prediction and cgMLST allele calling using genome assemblies (FASTA) |
Web & Standalone | https://lfz.corefacility.ca/sistr-app https://github.com/phac-nml/SISTR_cmd |
| SerotypeFinder | E. coli serotype prediction using raw sequencing reads (FASTQ) or genome assemblies (FASTA) |
Web & Standalone | https://cge.cbs.dtu.dk/services/SerotypeFinder |
| Kaptive | Capsule and lipopolysaccharide serotype prediction for Klebsiella pneumoniae and Acinetobacter baumannii genome assemblies (FASTA) |
Web & Standalone | https://kaptive-web.erc.monash.edu/ https://github.com/katholt/Kaptive |
| ClermonTyping | In silico Escherichia genus strain phylotyping using genome assemblies (FASTA) |
Web & Standalone | http://clermontyping.iame-research.center https://github.com/A-BN/ClermonTyping |
| pMLST | In silico plasmid multilocus sequence typing using genome assemblies (FASTA) |
Web & Standalone | https://pubmlst.org/organisms/plasmid-mlst |
| PlasmidFinder | In silico characterization of plasmid replicons from Enterobacteriaceae using raw sequencing reads (FASTQ) or genome assemblies (FASTA) |
Web & Standalone | https://cge.cbs.dtu.dk/services/PlasmidFinder |
| PLACNETw | A graph-based tool for plasmid reconstruction using raw sequencing reads (FASTQ) |
Web & Standalone | https://castillo.dicom.unican.es/upload https://github.com/LuisVielva/PLACNETw |
| oriTfinder | Identification of bacterial mobile genetic elements using genome assemblies (FASTA) or GenBank files |
Web & Standalone | https://bioinfo-mml.sjtu.edu.cn/oriTfinder |
| Phylogenetic inference | |||
| Gubbins | Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using a multi-FASTA alignment file |
Standalone | https://github.com/sanger-pathogens/gubbins |
| RecHMM | Detection of recombinant breakpoints and tree-topology changes in whole-genome sequence alignments using a multi-FASTA alignment file |
Standalone | http://biowiki.org/wiki/index.php/Rec_HMM |
| PhyML | Fast maximum likelihood-based phylogenetic inference using a PHYLIP format alignment file |
Web & Standalone | http://www.atgc-montpellier.fr/phyml https://github.com/stephaneguindon/phyml |
| RAxML | Phylogenetic analyses of large datasets under maximum likelihood using a multi-FASTA alignment file |
Standalone | https://github.com/stamatak/standard-RAxML |
| IQ-TREE | A fast and effective algorithm to infer phylogenetic trees by maximum likelihood using an alignment file in various common formats |
Web & Standalone | http://iqtree.cibiv.univie.ac.at http://www.iqtree.org |
| MrBayes | A variety of phylogenetic and evolutionary models based on Bayesian inference using a NEXUS format alignment file |
Standalone | http://nbisweden.github.io/MrBayes |
| BEAST | A cross-platform program for time-measured Bayesian phylogenetic analysis and phylodynamic data integration using an alignment file in various common formats |
Standalone | https://beast.community |
| Snippy | A pipeline for rapid variant calling and core genome alignment using raw sequencing reads (FASTQ) or genome assemblies (FASTA) |
Standalone | https://github.com/tseemann/snippy |
| Lyve-SET | A high-quality SNP pipeline to create a phylogeny using raw sequencing reads (FASTQ) |
Standalone | https://github.com/lskatz/lyve-SET |
| SNVPhyl | A pipeline for identifying high-quality SNPs and constructing a phylogenetic tree using raw sequencing reads (FASTQ) |
Standalone | https://snvphyl.readthedocs.io |
| CSI Phylogeny | Identifying variant and inferring phylogenies based on the concatenated alignment of the high-quality SNPs using raw sequencing reads (FASTQ) or genome assemblies (FASTA) |
Web | https://cge.cbs.dtu.dk/services/CSIPhylogeny |
| BacWGSTdb | A one-stop platform for bacterial whole-genome sequence typing and source tracking using genome assemblies (FASTA) |
Web | http://bacdb.cn/BacWGSTdb |
6.参考:
血清型_百度百科
汇总 | 基于全基因组测序数据的细菌分型方法
https://actamicro.ijournals.cn/actamicrocn/article/html/20220316
今天的分享到此就结束了,感谢您的阅读,如果确实帮到您,您可以动动手指转发给其他人。